Calculating summary statistics¶
This page is a tutorial on calculating summary statistics from a libsequence.VariantMatrix. We
also show how to interchange data between msprime [5] and pylibseq.
Simple overview of concepts¶
In [1]: import msprime
In [2]: import libsequence
In [3]: import numpy as np
# Get a TreeSequence from msprime
In [4]: ts = msprime.simulate(100, mutation_rate=250., recombination_rate=250., random_seed=42)
# TreeSequence -> VariantMatrix
In [5]: vm = libsequence.VariantMatrix.from_TreeSequence(ts)
# VariantMatrix -> AlleleCountMatrix
In [6]: ac = vm.count_alleles()
Certain summary statistics can be calculated simply from allele counts via
libsequence.AlleleCountMatrix. Calculations invovling linkage disequilibrium
will typically require the entire libsequence.VariantMatrix object.
The following examples rely on msprime [5] to generate input data, but the concepts hold more generally.
List of “classic”/standard summary statistics¶
Nucleotide diversity, or \(\pi\) [7]:
-
libsequence.thetapi(ac: libsequence._libsequence.AlleleCountMatrix) → float¶ Mean number of pairwise differences.
Parameters: ac – A libsequence.AlleleCountMatrixNote
Implemented as sum of site heterozygosity.
In [7]: print(libsequence.thetapi(ac))
980.3789898989783
Watterson’s estimator of \(\theta\), [10]:
-
libsequence.thetaw(ac: libsequence._libsequence.AlleleCountMatrix) → float¶ Watterson’s theta.
Parameters: m – A libsequence.AlleleCountMatrixNote
Calculated from the total number of mutations.
In [8]: print(libsequence.thetaw(ac))
982.5437651915445
Tajima’s \(D\), [8]:
-
libsequence.tajd(ac: libsequence._libsequence.AlleleCountMatrix) → float¶ Tajima’s D.
Parameters: m – A libsequence.AlleleCountMatrix
In [9]: print(libsequence.tajd(ac))
-0.007527225314052745
Fay and Wu’s test statistic, \(\pi - \hat\theta_H\) [2] has two overloads. The first takes a single ancestral state as an argument. The latter takes a list of ancestral states (one per site in the variant matrix):
-
libsequence.faywuh(*args, **kwargs)¶ Overloaded function.
- faywuh(ac: libsequence._libsequence.AlleleCountMatrix, ancestral_state: int) -> float
- faywuh(ac: libsequence._libsequence.AlleleCountMatrix, ancestral_states: List[int]) -> float
In [10]: print(libsequence.faywuh(ac, 0))
-74.02989898988403
The \(H'\) statistic [11] should be preferred to the Fay and Wu test statistic:
-
libsequence.hprime(*args, **kwargs)¶ Overloaded function.
- hprime(ac: libsequence._libsequence.AlleleCountMatrix, ancestral_state: int) -> float
- hprime(ac: libsequence._libsequence.AlleleCountMatrix, ancestral_state: List[int]) -> float
In [11]: print(libsequence.hprime(ac, 0))
-0.12962968409194914
Summaries of haplotype variation [1]:
-
libsequence.number_of_haplotypes(arg0: libsequence._libsequence.VariantMatrix) → int¶
-
libsequence.haplotype_diversity(arg0: libsequence._libsequence.VariantMatrix) → float¶
In [12]: print(libsequence.number_of_haplotypes(vm))
99
In [13]: print(libsequence.haplotype_diversity(vm))
���0.9997979797979798
The \(nS_L\) and \(iHS\) “haplotype homozygosity” statistics defined in [3]:
-
libsequence.nsl(m: libsequence._libsequence.VariantMatrix, refstate: int) → libsequence._libsequence.VecnSLResults¶
This function returns a list of instances of the following:
-
class
libsequence.nSLresults¶ Holds nSL, iHS, and non-reference count at core SNP
-
core_count¶ Core mutation count in sample
-
ihs¶ iHS
-
nsl¶ nSL
-
Values of nan imply that haplotype homozygosity for a particular core variant was not broken within the matrix. This is the same procedure used in [3]. The return value order corresponds to the row order of the input data:
In [14]: nsl = libsequence.nsl(vm, 0)
In [15]: for i in nsl[:4]:
....: print(i.core_count, i.ihs, i.nsl)
....:
1 nan nan
8 nan nan
3 nan nan
7 -0.09735588583687971 -0.5544227795580867
The following variant only allows haplotype homozygosity to be broken up by mutations where the non-reference count is \(\leq x\):
-
libsequence.nslx(m: libsequence._libsequence.VariantMatrix, refstate: int, x: int) → libsequence._libsequence.VecnSLResults¶
In [16]: nslx = libsequence.nslx(vm, 0, 1)
The haplotype diversity statistics from [4]:
-
libsequence.garud_statistics(arg0: libsequence._libsequence.VariantMatrix) → libsequence._libsequence.GarudStats¶
In [17]: g = libsequence.garud_statistics(vm)
In [18]: print(g.H1, g.H12, g.H2H1)
0.00020202020202020332 0.0006060606060606074 6.4401609045638545e-15
-
libsequence.two_locus_haplotype_counts(arg0: libsequence._libsequence.VariantMatrix, arg1: int, arg2: int, arg3: bool) → List[Sequence::TwoLocusCounts]¶
-
libsequence.allele_counts(arg0: libsequence._libsequence.AlleleCountMatrix) → List[libsequence._libsequence.AlleleCounts]¶
-
libsequence.non_reference_allele_counts(*args, **kwargs)¶ Overloaded function.
- non_reference_allele_counts(arg0: libsequence._libsequence.AlleleCountMatrix, arg1: int) -> List[libsequence._libsequence.AlleleCounts]
- non_reference_allele_counts(arg0: libsequence._libsequence.AlleleCountMatrix, arg1: List[int]) -> List[libsequence._libsequence.AlleleCounts]
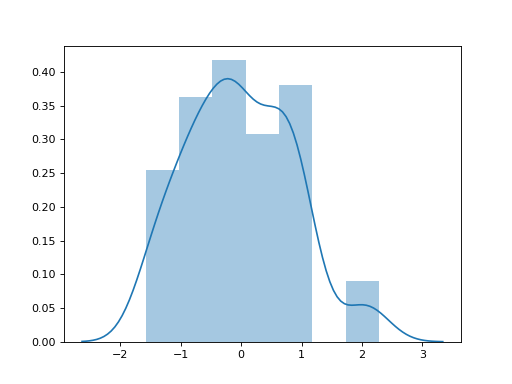
Distribution of Tajima’s D from msprime¶
import msprime
import libsequence
import seaborn as sns
D = []
for ts in msprime.simulate(100, num_replicates=100,
mutation_rate=25.0,
random_seed=42):
m = libsequence.VariantMatrix.from_TreeSequence(ts)
ac = m.count_alleles()
D.append(libsequence.tajd(ac))
sns.distplot(D)
“Sliding window” analysis¶
To analyze different genomic intervals, or “windows”, you have two options. Any statistic needing the allele count data can easily be gotten via a slice of your existing data:
In [19]: lefts = np.arange(0, 1, 0.2)
In [20]: for l in lefts:
....: p = np.where((vm.positions >= l)&(vm.positions < l+0.2))[0]
....: ac_slice=ac[p[0]:p[-1]+1]
....: assert len(ac_slice) == len(p)
....: print(libsequence.thetapi(ac_slice))
....:
179.54282828282925
198.07858585858628
230.90202020202207
196.7565656565662
175.0989898989905
We may also use libsequence.VariantMatrix.window() to obtain new VariantMatrix instances
corresponding to sub-windows of the data:
In [21]: for l in lefts:
....: w = vm.window(l, l+0.2)
....: acw = w.count_alleles()
....: print(libsequence.thetapi(acw))
....:
179.54282828282925
198.07858585858628
230.90202020202207
196.7565656565662
175.0989898989905
Window creation is \(O(log(vm.nsites))\) in time and has trivial additional memory requirements, as the returned object does not own its own data buffer.
Other useful statistics¶
Some more complex descriptors of the data are available
In [22]: diffs = libsequence.difference_matrix(vm)
Here, diffs contains data for representing the distance matrix for the data. Specifically, these values can be used to fill the upper triangle of a matrix:
In [23]: dm = np.array(diffs, dtype=np.int32)
In [24]: m = np.array([0]*(vm.nsam*vm.nsam))
# Get the indices of the upper-diagonal
# of an nsam*nsam matrix, ignoring
# the diagonal:
In [25]: idx = np.triu_indices(vm.nsam,k=1)
In [26]: m=m.reshape((vm.nsam,vm.nsam))
In [27]: m[idx]=dm
In [28]: print(m)
[[ 0 1053 940 ... 940 901 1017]
[ 0 0 1037 ... 1023 898 1038]
[ 0 0 0 ... 1102 1031 1101]
...
[ 0 0 0 ... 0 853 893]
[ 0 0 0 ... 0 0 1074]
[ 0 0 0 ... 0 0 0]]
# Let's confirm our results via brute-force:
In [29]: gm = ts.genotype_matrix()
In [30]: dummy = 0
In [31]: for i in range(gm.shape[1]-1):
....: for j in range(i+1,gm.shape[1]):
....: diffs = np.where(gm[:,i] != gm[:,j])
....: assert(len(diffs[0])==dm[dummy])
....: dummy+=1
....:
In a similar fashion, we can obtain true/false data on whether pairs of haplotypes differ:
In [32]: diff_yes_or_no = libsequence.is_different_matrix(vm)
-
libsequence.is_different_matrix(m: libsequence._libsequence.VariantMatrix) → List[int]¶ Return whether or not pairs of samples in a VariantMatrix differ
Parameters: m – A libsequence.VariantMatrix
The contents of this matrix have the exact same layout as diffs described above. The difference is that the data elements are encoded as 0 = identical, 1 = different. This calculation is much faster than the previous.
Note
Missing data do not contribute to samples being considered different, nor to the number of differences.
It is also possible to get a unique label assigned to each haplotype:
In [33]: labels = np.array(libsequence.label_haplotypes(vm),dtype=np.int32)
In [34]: print(len(np.unique(labels)))
99
-
libsequence.label_haplotypes(arg0: libsequence._libsequence.VariantMatrix) → List[int]¶
Internally, the results from is_different_matrix are used to assign the labels.
These labels are likewise used internally to count the number of haplotypes:
In [35]: print(libsequence.number_of_haplotypes(vm))
99
# Confirm result via direct comparison to
# the data from msprime:
In [36]: print(len(np.unique(gm.transpose(),axis=0)))
���99
What about performance?
In [37]: %timeit -n 10 -r 10 libsequence.number_of_haplotypes(vm)
10 loops, best of 10: 271 us per loop
In [38]: %timeit -n 10 -r 10 len(np.unique(gm.transpose(),axis=0))
10 loops, best of 10: 11.3 ms per loop