Methods dealing with recombination. More...
Functions | |
| double | HudsonsC (const Sequence::PolyTable *data, const bool &haveOutgroup, const unsigned &outgroup) __attribute__((deprecated)) |
| std::vector< PairwiseLDstats > | Disequilibrium (const Sequence::PolyTable *data, const bool &haveOutgroup=false, const unsigned &outgroup=0, const unsigned &mincount=1, const double max_distance=std::numeric_limits< double >::max()) __attribute__((deprecated)) |
| Calculate pairwise LD for a Sequence::PolyTable. More... | |
| PairwiseLDstats | PairwiseLD (const Sequence::PolyTable *data, unsigned i, unsigned j, const bool &haveOutgroup=false, const unsigned &outgroup=0, const unsigned &mincount=1, const double max_distance=std::numeric_limits< double >::max()) __attribute__((deprecated)) |
Variables | |
| const double | CMAX = 10000 |
Detailed Description
Methods dealing with recombination.
This namespace exists primarily so that the file Poly.cc (which defines Sequence::Poly) does not get too large. The routines defined in this namespace all have to do with properties of the association between sites. Current methods implemented are:
1.) Recombination::HudsonsC, which calculates Hudson's C, aka 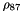
2.) Recombination::Disequilibrium, which calculated several measures of LD for all pairs of sites, and implements a frequency filter to remove low-frequency variants if desired.
Function Documentation
◆ Disequilibrium()
| std::vector< PairwiseLDstats > Sequence::Recombination::Disequilibrium | ( | const Sequence::PolyTable * | data, |
| const bool & | haveOutgroup = false, |
||
| const unsigned & | outgroup = 0, |
||
| const unsigned & | mincount = 1, |
||
| const double | max_distance = std::numeric_limits<double>::max() |
||
| ) |
Calculate pairwise LD for a Sequence::PolyTable.
This function is threaded (see threads) and implemented in terms of Sequence::PairwiseLD.
- Parameters
-
data A Sequence::PolyTable haveOutgroup A boolean outgroup. If haveOutgroup is true, then this is the index of the outgroup sequence in data mincount Minimum sample count to include a mutation. E.g., 2 = exclude singletons, etc. max_distance Do not include sites > this value apart
- Returns
- A std::vector of Sequence::PairwiseLDstats
Definition at line 353 of file Recombination.cc.
◆ HudsonsC()
| double Sequence::Recombination::HudsonsC | ( | const Sequence::PolyTable * | data, |
| const bool & | haveOutgroup, | ||
| const unsigned & | outgroup | ||
| ) |
Returns Hudson's (1987) Genetical Research 50:245-250 moment estimator of the population recombination rate. Please note that the properties of this estimator are not ideal, and one should prefer Hudson (2001) Genetics 159: 1805-1817 or McVean et al. (2002) Genetics 160: 1231-1241
Definition at line 329 of file Recombination.cc.
◆ PairwiseLD()
| PairwiseLDstats Sequence::Recombination::PairwiseLD | ( | const Sequence::PolyTable * | data, |
| unsigned | i, | ||
| unsigned | j, | ||
| const bool & | haveOutgroup = false, |
||
| const unsigned & | outgroup = 0, |
||
| const unsigned & | mincount = 1, |
||
| const double | max_distance = std::numeric_limits<double>::max() |
||
| ) |
Calculates LD statistics for sites i and j, where j>1.
- Parameters
-
data The polymorphism data i Index for site in data j Index for site in data haveOutgroup true if data contains an outgroup, false otherwise outgroup the index of the outgroup in data haveOutgroup is true mincount the minimum sample frequency to include max_distance max distance between markers (on scale in which positions in data are stored) for inclusion
- Returns
- Sequence::PairwiseLDstats
Definition at line 377 of file Recombination.cc.
Variable Documentation
◆ CMAX
| const double Sequence::Recombination::CMAX = 10000 |
The max value that can be taken by 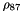
Definition at line 48 of file Recombination.cc.
