9. Gene dropping on pedigrees#
This chapter goes into more detail on how we go from a pedigree to a gene tree. We build on the discussion in Section 3.
Fig. 9.1 shows a pedigree with two siblings.
This pedigree also shows the alleles present in the parents.
As in Section 3, we assume autosomal inheritance in diploids.
We label our parents with four distinct alleles, 1, 2, 3, and 4.
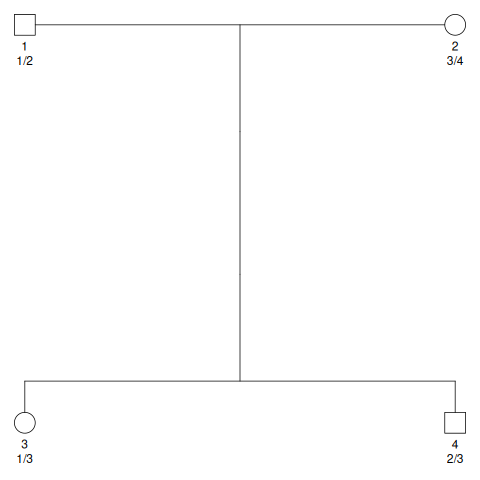
Fig. 9.1 A pedigree with two siblings. Under each individual label, the genotype is shown for an autosomal locus. The father (individual 1) is shown as a heterozygote for alleles 1 and 2. The mother (individual 2) is shown as a heterozygote for alleles 3 and 4. The offspring genotypes show which parental alleles were inherited.#
In order to turn the pattern of allele transmission in our pedigree into a gene tree, we follow the convention stated in Section 3, which is repeated here:
For an individual in the pedigree with label \(i\), that individual’s two genomes will be labelled \(2i - 1\) and \(2i\)
The reason why we do this is that we need to associate a node (which is a genome or allele) with a birth time.
Our parental alleles already follow our convention. But for our offspring, we will require that:
Individual 3 corresponds to alleles 5 and 6.
Individual 4 corresponds to alleles 7 and 8.
(Remember – two alleles in a diploid for an autosomal gene!)
From Fig. 9.1, we see that individual 3 inherited alleles 1 and 3. We will relabel those as 5 and 6. Individual 4 inherited alleles 2 and 3, which we will relabel to 7 and 8.
After relabelling, we can now draw our gene tree (Fig. 9.2).
Fig. 9.2 An example of gene dropping and labelling alleles. The image is based on the pedigree shown in Fig. 9.1. For alleles found in the pedigree children, the original allele labels from Fig. 9.1 are shown in parentheses.#