Class List
Here are the classes, structs, unions and interfaces with brief descriptions:
[detail level 123]
| ▼NSequence | The namespace in which this library resides |
| ▼Ncoalsim | Routines for coalescent simulation |
| Cchromosome | A chromosome is a container of segments |
| Cmarginal | The genealogy of a portion of a chromosome on which no recombination has occurred |
| Cnewick_stream_marginal_tree | Class that provides a typecast-on-output of a marginal tree to a newick tree Example use: |
| Cnewick_stream_marginal_tree_impl | |
| Cnode | A point on a marginal tree at which a coalescent event occurs |
| Csegment | A portion of a recombining chromosome |
| ▼Ninternal | |
| Ccol_view_ | Implementation details for Sequence::ColView and Sequence::ConstColView |
| Crow_view_ | Implementation details for Sequence::RowView and Sequence::ConstRowView |
| C_PolySNPImpl | |
| CAlignStream | Virtual interface to alignment streams |
| CAlleleCountMatrix | Matrix representation of allele counts in a VariantMatrix To be constructed |
| CAlleleCounts | |
| CambiguousNucleotide | Tests if a character is in the set A,G,C,T |
| CClustalW | ClustalW streams |
| Ccol_view_iterator | Iterator for column views |
| CComeron95 | Ka and Ks by Comeron's (1995) method |
| CComplementBase | |
| CcountDerivedStates | Functor to count the number of derived states, excluding gaps and missing data, in a range of characters |
| CFasta | FASTA sequence stream |
| Cfastq | |
| CFST | Analysis of population structure using 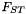 |
| CGarudStats | |
| CGrantham | Grantham's distances |
| CGranthamWeights2 | Weights paths by Grantham's distances for codons differing at 2 sites |
| CGranthamWeights3 | Weights paths by Grantham's distances for codons differing at 3 sites |
| CHKAdata | Data from a single locus for an HKA test |
| CHKAresults | Results of calculations of the HKA test |
| CinvalidPolyChar | This functor can be used to determine if a range contains characters that the SNP analysis routines in this library cannot handle gracefully |
| CKimura80 | Kimura's 2-parameter distance |
| Cnmuts | Calculate the number of mutations at a polymorphic site |
| CnSLiHS | |
| CPairwiseLDstats | Pairwise linkage disequilibrium (LD) stats |
| CphylipData | |
| CPolySIM | Analysis of coalescent simulation data |
| CPolySites | Polymorphism tables for sequence data |
| CPolySNP | Molecular population genetic analysis |
| CPolyTable | The base class for polymorphism tables |
| CPolyTableSlice | A container class for "sliding windows" along a polymorphism table |
| CRedundancyCom95 | Calculate redundancy of a genetic code using Comeron's counting scheme |
| CSeq | Abstract interface to sequence objects |
| CshortestPath | Calculate shortest path between 2 codons |
| CSimData | Data from coalescent simulations |
| CSimParams | Parameters for Hudson's simulation program |
| CSimpleSNP | SNP table data format |
| CSingleSub | Deal with codons differing at 1 position |
| CSites | Calculate length statistics for divergence calculations |
| CstateCounter | Keep track of state counts at a site in an alignment or along a sequence |
| CStateCounts | Track character state occurrence at a site in a VariantMatrix |
| CSums | |
| CThreeSubs | Deal with codons differing at all 3 positions |
| CTwoSubs | Deal with codons differing at 2 positions |
| CUnweighted2 | Weights all pathways equally |
| CUnweighted3 | Weights all pathways equally |
| CVariantMatrix | Matrix representation of variation data |
| CWeightingScheme2 | Abstract interface to weighting schemes when codons differ at 2 positions |
| CWeightingScheme3 | Abstract interface to weighting schemes when codons differ at 3 positions |
| Cfasta_constructors_fixture | \ file FastaConstructors.cc |
| Cfirst_is_equal | Functor that checks for equality of first member of two pairs |
| CoutputFreq | |
| Cssh | Calculate nucleotide diversity from a polymorphic site |
