Timing of events in demographic models#
This document provides several examples showing when events happen in demographic models.
We will define very simple setups that allow very simple assertions about their outcomes. While most simulations will involve more complex setups, the advantage of simplicity here is that we can easily validate the results because we can easily reason about the models.
These examples come from the fwdpy11 test suite.
The key concept here is that the demes specification
defines an epoch as a half open interval on [present, past).
Our reasoning about the outcomes of data that we record during simulation involves breaking our models up into half open intervals and asking questions about them.
Why do we spend time going over this? Imagine that you want to know the details about the ancestry of a given generation in your simulation. For example, the ancestry of the generation born during a pulse. One way to do that would be to preserve both the pulse generation and its parent generation as ancient samples. (See here.) To do such sampling you need to know when to sample!
Some setup#
During some of our simulations, we will record the current time point and the part of the metadata containing individual parents.
We can use a simple data class to record this information:
@dataclass
class Parents:
generation: int
parents: np.array
@dataclass
class RecordParents:
parents: list
def __call__(self, pop, _):
md = np.array(pop.diploid_metadata, copy=False)
self.parents.append(Parents(pop.generation, np.copy(md["parents"])))
There will be no genetics happening during the simulation. All that happens is that we apply our recorder. We will always “burn in” the demographic model with 100 generations of matings.
def run_model(yaml, recorder):
graph = demes.load(yaml)
demog = fwdpy11.ForwardDemesGraph.from_demes(
graph, burnin=100, burnin_is_exact=True
)
pdict = {
"nregions": [],
"recregions": [],
"sregions": [],
"rates": (0.0, 0.0, 0.0),
"gvalue": fwdpy11.Multiplicative(scaling=2.0),
"simlen": demog.final_generation,
"demography": demog,
}
params = fwdpy11.ModelParams(**pdict)
rng = fwdpy11.GSLrng(101)
pop = fwdpy11.DiploidPopulation(demog.initial_sizes, 1)
fwdpy11.evolvets(rng, pop, params, simplification_interval=100, recorder=recorder)
return pop
Intervals when demes exist#
In the following graph, demes completely replace one another over time:
time_units: generations
demes:
- name: deme0
epochs:
- end_time: 50
start_size: 100
- name: deme1
ancestors: [deme0]
epochs:
- end_time: 25
start_size: 100
- name: deme2
ancestors: [deme1]
epochs:
- start_size: 100
The following graphic depicts the model:
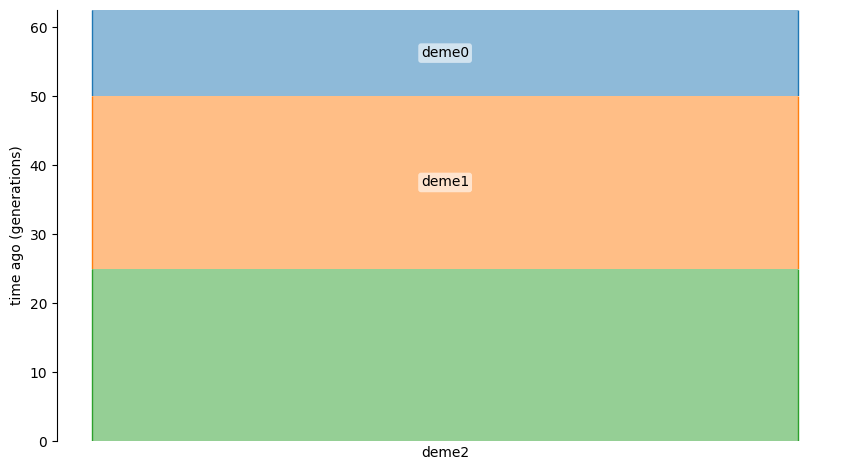
The intervals of a deme’s existence are from \([i, j)\) generations ago, where \(j > i\).
Therefore deme2 exists from \([0, 25)\) generations ago, etc..
During simulation, a node’s birth time and deme are recorded. Therefore, we can export the simulation to a tree sequence and check that the node times match up with their expected deme labels:
yaml = make_path("demes_event_examples/deme_existence.yaml")
recorder = None
pop = run_model(yaml, recorder)
assert pop.generation == 150
ts = pop.dump_tables_to_tskit()
assert all([i.population == 2 for i in ts.nodes() if i.time < 25.0])
assert all(
[i.population == 1 for i in ts.nodes() if i.time >= 25.0 and i.time < 50]
)
assert all([i.population == 0 for i in ts.nodes() if i.time >= 50])
A single pulse event#
The following graph defines a two deme model with a single pulse event 10 generations ago.
time_units: generations
demes:
- name: deme0
epochs:
- start_size: 100
- name: deme1
epochs:
- start_size: 100
pulses:
- sources: [deme0]
dest: deme1
proportions: [1.0]
time: 10
The following graphic depicts the model:
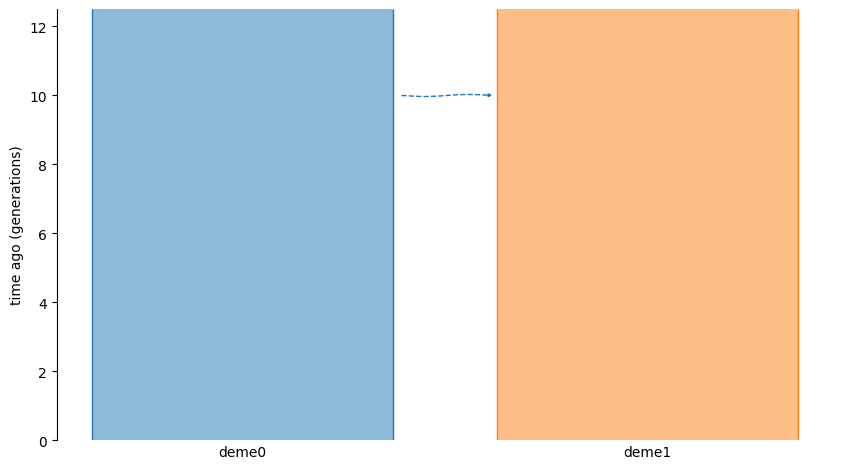
Given this model:
The time interval \([0, 10)\) generations ago is the post pulse interval (thinking forwards in time). During this interval, all individuals in
deme0have parents from that same deme. Likewise fordeme1.All individuals born in the pulse generation (10 generations ago) have all of their parents drawn from
deme0. The reason for this is that there are no events affecting the ancestry ofdeme0and the proportion ofdeme1ancestry that comes fromdeme0is 1.0, which is all of it.The time interval \([11, \infty)\) generations ago is the pre pulse interval (thinking forwards in time). During this interval, all individuals in
deme0have parents from that same deme. Likewise fordeme1.
Using the reasoning about the pre and post epochs with respect to the timing of the pulse, we can make the following assertions:
yaml = make_path("demes_event_examples/single_pulse.yaml")
recorder = RecordParents([])
pop = run_model(yaml, recorder)
assert pop.generation == 110
assert (
np.all(p.parents.flatten() < 100)
for p in recorder.parents
if p.generation == pop.generation - 10
)
for p in recorder.parents:
if p.generation != pop.generation - 10:
flat = p.parents.flatten()
deme0 = flat[np.where(flat < 100)]
deme1 = flat[np.where(flat >= 100)]
assert len(deme0) == 200, f"{p}"
assert len(deme1) == 200, f"{p}"
Let’s work through the logic behind assertions in detail:
The size of each deme is 100 diploids.
The metadata are ordered by deme id. Thus, the first 100 entries are the indexes parents of individuals in
deme0, etc..Due to the metadata ordering, an index \(< 100\) (or, \([0, 100)\)) indicates a parent from
deme0. An index in \([100, 200)\) indicates a parent fromdeme1.
A multi-generation “burst” of migration#
This model is a slight twist on the previous. Rather than a single generation pulse, there are several consecutive generations of migration in one direction:
time_units: generations
demes:
- name: deme0
epochs:
- start_size: 100
- name: deme1
epochs:
- start_size: 100
migrations:
- source: deme0
dest: deme1
rate: 1.0
start_time: 10
end_time: 5
The following graphic depicts the model:
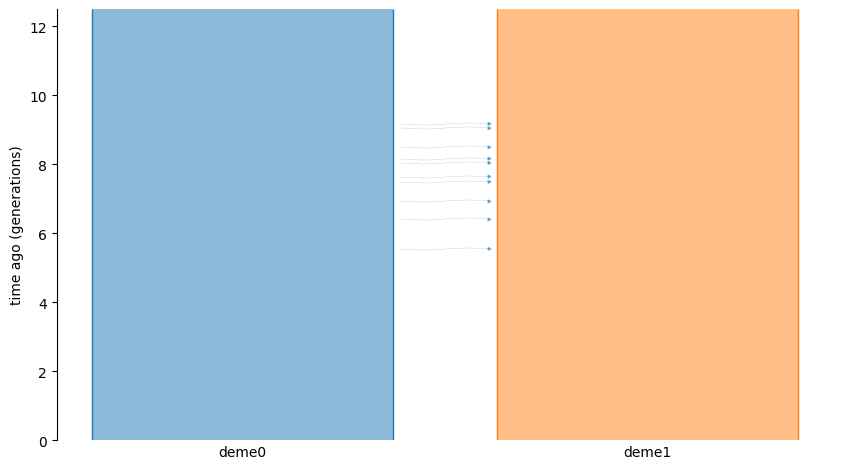
In this model, migration occurs during the interval \([5, 10)\) generations ago.
Because the migration is one way (deme0 to deme1) and the rate is 1.0, the parents
of all offspring born in this interval have their parents sampled from deme0.
Outside of this interval, the parents of any individual are drawn from that individual’s deme.
The following code asserts that these intervals are what we expect:
yaml = make_path("demes_event_examples/burst_of_migration.yaml")
recorder = RecordParents([])
pop = run_model(yaml, recorder)
assert pop.generation == 110
assert (
np.all(p.parents.flatten() < 100)
for p in recorder.parents
if p.generation > pop.generation - 10 and p.generation <= pop.generation - 5
)
for p in recorder.parents:
if p.generation <= pop.generation - 10 or p.generation > pop.generation - 5:
flat = p.parents.flatten()
deme0 = flat[np.where(flat < 100)]
deme1 = flat[np.where(flat >= 100)]
assert len(deme0) == 200, f"{p}"
assert len(deme1) == 200, f"{p}"