Tracking mutations over time using sqlite3#
Note
This is an advanced vignette. It shows off how powerful it is to have access to the entire Python world of data packages during a simulation. This vignette uses low-level sqlite syntax as well as higher-level Python libraries.
Introduction#
The previous vignette tracking mutation frequencies was straightforward to implement.
However, using dict to key lists of frequencies risks running out of memory.
This vignette reimplements the previous method using much more powerful techniques:
Instead of the
dict, we record data into fixed-lengthnumpy.ndarrayobjects.During the simulation, we preserve all individuals of each generation as ancient samples.
See here for background. The trick is that we will only keep these ancient samples until the table collection is simplified!We populate those arrays immediately after simplification. We use the
post_simplification_recorderargument tofwdpy11.evolvets(), passing in a class that gets frequencies from the tree sequences. Once our callback is finished, the ancient samples are purged.When the
numpyarrays get full, we transfer their data to a relational database using thesqlite3module that is built in to the Python language. We will take advantage of thepandas.DataFramefunctionpandas.DataFrame.to_sql(), which can transfer in-memory data toSQLdatabases via a convenientAPI.
We will need the following imports:
import attr
import sqlite3
import fwdpy11
import numpy as np
import pandas as pd
We need to implement two Python classes. The first will handle the mechanics of recording the ancient samples themselves. This class is trivial (see here for background):
Helper types#
class SampleEveryone(object):
def __call__(self, pop, sampler):
sampler.assign(np.arange(pop.N, dtype=np.uint32))
The second class will be a callable that gets executed immediately after simplification.
This type is responsible for processing trees, getting data into numpy arrays, and transferring the data to the database.
This class has a lot of responsibilities.
In order to shorten the code, we define a TableBuffer class that holds the numpy arrays and handles the mechanics of calling pandas.DataFrame.to_sql().
The definition of this class is shown below, and you may click to see it.
Show code cell source
@attr.s(auto_attribs=True, kw_only=True)
class TableBuffer(object):
max_records: int
def __attrs_post_init__(self):
self.num_records = 0
self.generation = np.zeros(self.max_records, dtype=np.uint32)
self.origin_time = np.zeros(self.max_records, dtype=np.uint32)
self.position = np.zeros(self.max_records, dtype=np.float64)
self.frequency = np.zeros(self.max_records, dtype=np.float64)
self.effect_size = np.zeros(self.max_records, dtype=np.float64)
def update(
self,
conn,
*,
generation,
origin_time,
position,
frequency,
effect_size,
):
self.generation[self.num_records] = generation
self.origin_time[self.num_records] = origin_time
self.position[self.num_records] = position
self.frequency[self.num_records] = frequency
self.effect_size[self.num_records] = effect_size
self.num_records += 1
if self.num_records == self.max_records:
self._flush_buffer(conn)
def _flush_buffer(self, conn):
if self.num_records == 0:
return
df = pd.DataFrame(
{
"generation": self.generation[: self.num_records],
"origin_time": self.origin_time[: self.num_records],
"position": self.position[: self.num_records],
"frequency": self.frequency[: self.num_records],
"effect_size": self.effect_size[: self.num_records],
}
)
df.to_sql(
"trajectories_temp",
conn,
if_exists="append",
index=False,
method="multi",
chunksize=25000,
)
self.num_records = 0
We also have two simple helper classes. We will use them to not record the frequencies of fixed mutations over and over again, as well as to record when a mutation did reach fixation.
@attr.s(auto_attribs=True, kw_only=True, frozen=True)
class MutationKey(object):
origin_time: int
position: float
effect_size: float
@attr.s(auto_attribs=True, kw_only=True, frozen=True)
class FixationRecord(object):
fixation_time: int
A class to track mutations frequencies via a database#
Now, we define CountMutations, which is our real workhorse.
The __call__ function takes an instance of fwdpy11.DiploidPopulation as an argument.
When this function is called, the fwdpy11.DiploidPopulation.tables will be indexed, allowing us to visit trees using fwdpy11.TreeIterator.
This class also contains several “boiler plate” functions to handle data transfer to the database. We follow the Python tradition of using function names beginning with an underscore to indicate that these functions are not intended to be part of the public interface to this class.
Note
This example uses an in-memory SQL database named :memory:.
(See sqlite3.connect() for details.)
We use this approach because this example may be run on a cloud server.
In practice, it is much more efficient to use an on-disk database!
Doing so requires some simple modifications that are left as an “exercise for the reader”.
On a first read, you probably want to skip all of the underscore functions and focus on the details of __call__.
This function uses fwdpy11.DiploidPopulation.sample_timepoints() to iterate over the ancient samples based on their birth times.
For each cohort of ancient samples, we simplify the tables with respect to that sample using fwdpy11.simplify_tables().
We then iterate over all the trees, using fwdpy11.TreeIterator, to obtain the number of samples with each mutation via fwdpy11.TreeIterator.leaf_counts().
The implementation details of __call__ are offloaded to another function.
The reason is that we need to avoid double-counting mutations!
When the tables are simplified, they are simplified with respect to the ancestry of all samples nodes, both “alive” and “ancient”.
Because we are preserving every generation, our current alive nodes will soon become ancient samples.
Thus, to avoid double counting, we do not process the currently-alive generation.
To make sure that we do, eventually, get all mutations counted, we need to call our count_em_up function at the end of the simulation for final tallying.
@attr.s(auto_attribs=True)
class CountMutations(object):
"""
This is run only in generations where simplification happens.
Tables are indexed and "ready to roll" for analysis.
"""
max_records: int
def __attrs_post_init__(self):
self.buffer = TableBuffer(max_records=self.max_records)
self.num_records = 0
self.fixations: typing.Dict = dict()
self.conn = sqlite3.connect(":memory:")
def _dump_fixations(self):
origin_time = []
effect_size = []
position = []
fixation_times = []
for k, v in self.fixations.items():
origin_time.append(k.origin_time)
effect_size.append(k.effect_size)
position.append(k.position)
fixation_times.append(v.fixation_time)
df = pd.DataFrame(
{
"origin_time": origin_time,
"effect_size": effect_size,
"position": position,
"fixation_time": fixation_times,
}
)
df.to_sql("fixations_temp", self.conn, if_exists="replace", index=False)
self.fixations = dict()
def _dump_remaining_data(self):
self.buffer._flush_buffer(self.conn)
self._dump_fixations()
def finalize_database(self):
"""
Be sure to call this at the end of a simulation!
"""
self._dump_remaining_data()
self.conn.execute(
"create table mutations (origin_time int, position real, effect_size real, id integer primary key autoincrement)"
)
self.conn.commit()
self.conn.execute(
"""
insert into mutations select origin_time, position, effect_size, NULL
from
trajectories_temp
group by origin_time, position, effect_size
"""
)
self.conn.commit()
self.conn.execute(
"""
create table trajectories
as
select mutations.id, trajectories_temp.generation,
trajectories_temp.frequency
from
trajectories_temp
left join mutations
on trajectories_temp.origin_time == mutations.origin_time
and trajectories_temp.position == mutations.position
and trajectories_temp.effect_size == mutations.effect_size
"""
)
self.conn.execute("drop table trajectories_temp")
self.conn.execute(
"""
create table fixations
as
select mutations.id, fixations_temp.fixation_time
from fixations_temp
left join mutations
on fixations_temp.origin_time == mutations.origin_time
and fixations_temp.position == mutations.position
and fixations_temp.effect_size == mutations.effect_size
"""
)
self.conn.execute("drop table fixations_temp")
self.conn.commit()
# indexes speed up cross-table analysis
self.conn.execute("create index traj_id_index on trajectories(id)")
self.conn.execute("create index fixation_id_index on fixations(id)")
self.conn.execute("create index mutation_id_index on mutations(id)")
self.conn.execute("vacuum")
self.conn.commit()
def count_em_up(self, pop, *, include_alive):
"""
We implement the mechanics of __call__ separately,
so that they may be re-used to process alive nodes
when evolvets returns.
"""
for generation, nodes, metadata in pop.sample_timepoints(
include_alive=include_alive
):
# non-owning view
w = metadata["w"][:]
tables, idmap = fwdpy11.simplify_tables(pop.tables, nodes)
tv = fwdpy11.TreeIterator(tables, idmap[nodes])
for t in tv:
for m in t.mutations():
key = MutationKey(
origin_time=pop.mutations[m.key].g,
position=pop.mutations[m.key].pos,
effect_size=pop.mutations[m.key].s,
)
if key not in self.fixations:
s = t.leaf_counts(m.node)
assert s > 0
freq = s / (2.0 * pop.N)
if freq == 1.0:
self.fixations[key] = FixationRecord(
fixation_time=generation
)
self.buffer.update(
self.conn,
generation=generation,
origin_time=key.origin_time,
position=key.position,
frequency=freq,
effect_size=key.effect_size,
)
def __call__(self, pop):
self.count_em_up(pop, include_alive=False)
We now run the same simulation as in in the previous vignette. The differences are:
An instance of
SampleEveryoneis our recorder.An instance of
CountMutationsis passed topost_simplification_recorderWe simplify every 100 generations instead of every generation.
pdict = {
# Add a region for neutral mutations:
"nregions": [fwdpy11.Region(0, 1, 1)],
"sregions": [fwdpy11.ExpS(beg=0, end=1, weight=1, mean=0.2)],
"recregions": [fwdpy11.PoissonInterval(0, 1, 1e-2)],
"gvalue": [fwdpy11.Multiplicative(2.0)],
"demography": fwdpy11.ForwardDemesGraph.tubes([100], burnin=100, burnin_is_exact=True),
# Add a mutation rate for neutral mutations:
"rates": (5e-3, 1e-3, None),
"simlen": 100,
}
params = fwdpy11.ModelParams(**pdict)
pop = fwdpy11.DiploidPopulation(100, 1.0)
rng = fwdpy11.GSLrng(54321)
recorder = SampleEveryone()
counter=CountMutations(max_records=100)
fwdpy11.evolvets(
rng,
pop,
params,
100, # Simplify every 100 generations
recorder,
suppress_table_indexing=False,
post_simplification_recorder=counter
)
As mentioned earlier, we have to run a couple of extra functions now to make sure we process all of our data:
# Get data from the final generation
counter.count_em_up(pop, include_alive=True)
counter.finalize_database()
We are going to use pandas.read_sql() to pull data from our database into pandas.DataFrame objects.
Our approach here is not optimal and we discuss alternatives below.
traj = pd.read_sql('select * from trajectories', counter.conn)
mut = pd.read_sql('select * from mutations', counter.conn)
df = pd.merge(traj, mut, how='left', on=['id'])
df['neutral'] = [i == 0.0 for i in df.effect_size]
Finally, let us plot our output:
import matplotlib.pyplot as plt
f, ax = plt.subplots()
for n, g in df.groupby(['id', 'neutral']):
if len(g.index) >= 10:
linestyle = '-'
alpha=1.0
if n[1] is True:
linestyle='--'
else:
alpha=0.5
ax.plot(g.generation, g.frequency, linestyle=linestyle, alpha=alpha)
ax.set_xlabel("Time (generation)")
ax.set_ylabel("Frequency");
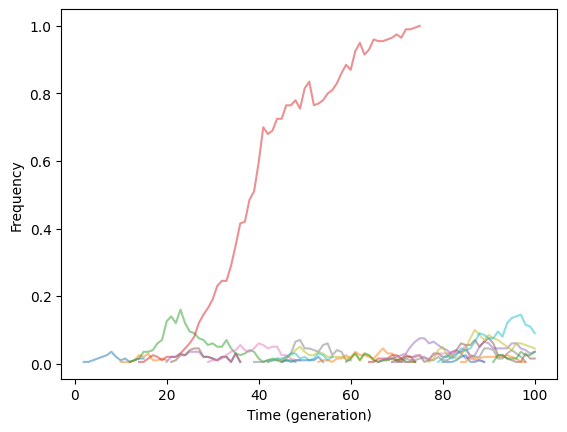
The figure should be identical to Fig. 2.
Details of the implementation#
What are all the underscore methods doing?
They are all called by finalize_database, which is to be run at the end of a simulation.
The operations are:
Create a
mutationstable in our database. We populate this table based on unique(position, effect size, origin time)combinations in our temporary trajectories. This table has an automatically incrementing column that assigns a unique integerIDto each combination. This unique integer will be a “mutationID”.We can now create our final trajectories table that is populated with the mutation
ID, generation in the simulation and frequency. Thus, the redundant information is removed.We create a
fixationstable that also uses theseIDvalues.Finally, we create index columns for the
IDvalues for each table. These indexes will greatly speed up analysis.
The use of all of this is to both save space on disk and to speed up analysis.
Further considerations#
Like the previous vignette, this recipe needs adjustment for the following cases:
Tracking mutation frequencies with respect to deme.
Tracking mutation frequencies in demes that change size.
When processing the databases, it is best to do as much as possible at the level of
SQLitself and not pull everything into a data frame like we did above. For on-disk databases,R’s dbplyr package will generally be the best tool here. Yes, this means changing languages. But, you get to use the same syntax as “regular” dplyr for most of your analyses. Most importantly, by havingdbplyrexecute your analysis on theSQLside, you can analyze massive data sets that you could not possible hold in memory. You can plot the output usingRor use any of a variety of methods to get your processed data back into Python for plotting.