Using demes to specify demographic models.#
Note
The lists of objects described here are passed to the demography and simlen parameters when initializing instances of fwdpy11.ModelParams.
Starting with version 0.14.0, it is possible to specify a demographic model using the demes specification.
This method of specifying models uses a YAML syntax, which is described in detail in the demes specification.
It is a good idea to browse both the tutorial and the gallery before continuing.
There are several advantages to using demes:
The
YAMLfiles can be used by many pieces of related software, includingmsprimeandmoments.You’ll get an extra layer of model validation when
demesloads your model prior to it being converted intofwpdy11objects.Tools to visualize the models are under active development.
YAML input#
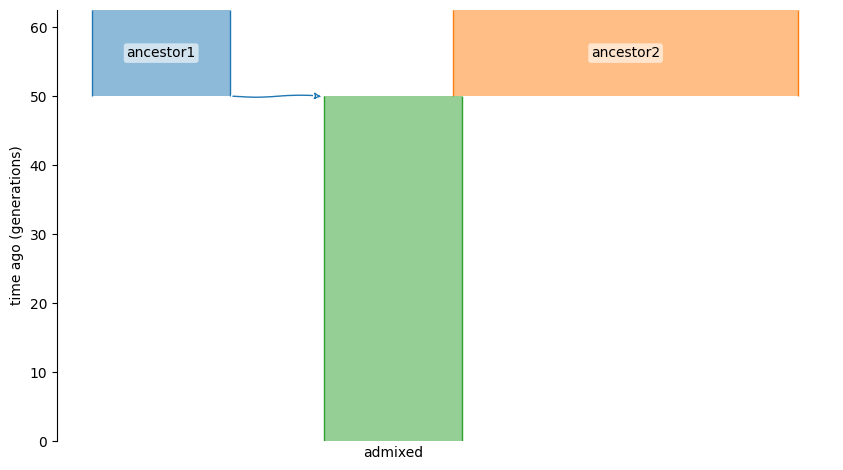
The following YAML specifies a model of a derived population created
from unequal admixture of two ancestral populations 50 generations ago:
yaml="""
description: An example demes model
time_units: generations
demes:
- name: ancestor1
epochs:
- start_size: 100
end_time: 50
- name: ancestor2
epochs:
- start_size: 250
end_time: 50
- name: admixed
start_time: 50
ancestors: [ancestor1, ancestor2]
proportions: [0.90, 0.10]
epochs:
- start_size: 100
"""
A visual representation of the model looks like:
Show code cell source
import demes
import demesdraw
graph = demes.loads(yaml)
demesdraw.tubes(graph);
We can generate demographic models directly from these YAML files using
fwdpy11.ForwardDemesGraph.from_demes().
import fwdpy11
demography = fwdpy11.ForwardDemesGraph.from_demes(yaml, burnin=100, burnin_is_exact=True)
The previous command defines a model where we evolve the population for 100 generations
of a “burn-in”.
When burnin_is_exact=False, the value passed to burnin is treated as a multiple
of the sum of all ancestral deme sizes (those having a start time of inf
in the demes model).
Note
Be sure to read the documentation for fwdpy11.ForwardDemesGraph.from_demes()!
There are important options concerning the run time of the simulation, etc.
The return value is an instance of fwdpy11.ForwardDemesGraph.
The object contains several properties that are useful in setting up your
simulation.
First, the sizes of all demes extant at generation zero:
demography.initial_sizes
[100, 250]
This property can be used to correctly initialize a population:
pop = fwdpy11.DiploidPopulation(demography.initial_sizes, 1000.)
print(pop.deme_sizes())
(array([0, 1], dtype=int32), array([100, 250]))
The generation corresponding to the end of the model:
demography.final_generation
150
We can now set up our simulation parameters:
pdict = {'simlen': demography.final_generation,
'demography': demography,
# Everything below is standard
# and covered in other vignettes
'recregions': [],
'nregions': [],
'sregions': [],
'rates': (0,0,0),
'gvalue': fwdpy11.Multiplicative(2.)
}
params = fwdpy11.ModelParams(**pdict)
We are now ready to run a simulaton:
We have a valid
fwdpy11.ModelParamsobject.Our population has its initial sizes correctly set.
Obtaining the demes present at the end of a simulation#
demography.demes_at_final_generation
[2]
To get the names of those demes:
for deme in demography.demes_at_final_generation:
print(demography.deme_labels[deme])
admixed
It is probably useful to read more about how fwdpy11 handles demes
models here.
More information#
See here for concrete examples about reasoning through when demographic events occur.