File List
Here is a list of all documented files with brief descriptions:
| algorithm.hpp | |
| Alignment.hpp | Declaration of namespace Sequence::Alignment |
| Alignment.tcc | |
| AlignmentTest.cc | Unit tests for fxns in namespace Sequence::Alignment |
| AlignStream.hpp | Declaration of virtual base class for alignment streams (Sequence::AlignStream) |
| AlignStream.tcc | |
| AlignStreamTest.cc | Unit tests for Sequence::ClustalW and Sequence::phylipData |
| allele_counts.cc | |
| allele_counts.hpp | Count alleles at variable sites |
| AlleleCountMatrix.cc | |
| AlleleCountMatrix.hpp | |
| alphabets.cc | Unit tests for Sequence/SeqAlphabets.hpp |
| auxillary.cc | |
| auxillary.hpp | |
| baseComp.cc | |
| classics.hpp | "Classic" summaries of variation data |
| Clustalw.hpp | Declaration of Sequence::ClustalW streams |
| Clustalw.tcc | |
| Coalesce.hpp | |
| Coalesce.tcc | |
| Coalescent.hpp | A lazy header to include the headers needed to start writing simulations. Includes: <Sequence/Coalescent/SimTypes.hpp> <Sequence/Coalescent/Coalesce.hpp> <Sequence/Coalescent/Recombination.hpp> <Sequence/Coalescent/Mutation.hpp> <Sequence/Coalescent/Initialize.hpp> <Sequence/Coalescent/DemographicModels.hpp> <Sequence/Coalescent/FragmentsRescaling.hpp> |
| CoalescentCoalesce.cc | |
| CoalescentFragmentsRescaling.cc | |
| CoalescentInitialize.cc | |
| CoalescentMutation.cc | |
| CoalescentRecombination.cc | |
| CoalescentSimTypes.cc | |
| CoalescentTreeOperations.cc | |
| codons.cc | |
| CodonTable.cc | |
| CodonTable.hpp | Facility to count codons in CDS sequence, function Sequence::makeCodonUsageTable |
| col_view_iterator.hpp | |
| Comeron95.cc | |
| Comeron95.hpp | Declaration of Sequence::Comeron95 to calculate Ka/Ks |
| Comparisons.cc | |
| Comparisons.hpp | Delcaration of routines for comparing DNA sequences This file declares a set of functions useful for comparing two bits of sequence data–sequences, nucleotides, etc |
| ComparisonsTest.cc | |
| ComplementBase.cc | |
| ComplementBase.hpp | Delcaration of Sequence::ComplementBase, a function object to return the complement of a DNA nucleotide |
| config.h | |
| correlations.cc | |
| CountingOperators.cc | |
| CountingOperators.hpp | Declarations of operators to add associative containers together |
| CountingOperators.tcc | |
| DemographicModels.hpp | |
| DemographicModels.tcc | |
| descriptiveStats.hpp | Classes and functions for calculating descriptive statistics of lists of random variables |
| descriptiveStats.tcc | |
| Fasta.cc | |
| Fasta.hpp | Declaration of Sequence::Fasta streams |
| FastaConstructors.cc | |
| FastaExplicitIO.cc | |
| FastaIO.cc | |
| FastaOperations.cc | |
| fastq.cc | |
| fastq.hpp | FASTQ class |
| fastqConstructors.cc | |
| fastqIO.cc | |
| faywuh.cc | |
| filtering.cc | |
| filtering.hpp | |
| FragmentsRescaling.hpp | Helper functions for simulating partially linked fragments One often wants to simulate partially linked fragments under neutral models. An efficient way to do this is to simulate a contiguous fragment, but with the recombination rate varying along the region (to represent the variable genetic distances between fragments). This header file declares functions that make this task easier, particularly the operations of rescaling the positions of mutations/marginal trees from the genetic map back to the physical map |
| fst.cc | |
| FST.cc | |
| FST.hpp | Delcaration of a class (Sequence::FST) to analyze population structure |
| garud.cc | |
| Garud.cc | |
| garud.hpp | H1, H12, and H2/H1 stats |
| Garud.hpp | |
| generic.cc | |
| generic.hpp | Generic utilities for calculating summary statistics |
| Grantham.cc | |
| Grantham.hpp | Grantham's distances (Sequence::Grantham) |
| GranthamWeights.cc | |
| GranthamWeights.hpp | Declaration of classes to weight codons by Grantham distance (i.e. for Sequence::Comeron95). Declares Sequence::GranthamWeights2 and Sequence::GranthamWeights3 |
| haplotype_statistics.cc | |
| HKA.cc | |
| HKA.hpp | Calculations related to the HKA test |
| hprime.cc | |
| hprime_faywuh_aggregator.hpp | |
| Hudson2001.hpp | |
| Initialize.hpp | |
| int_handler.cc | |
| int_handler.hpp | |
| Kimura80.cc | |
| Kimura80.hpp | Declaration of Sequence::Kimura80 |
| ld.cc | |
| ld.hpp | |
| lHaf.cc | |
| lHaf.hpp | |
| libseq_unit_tests.cc | |
| libsequenceConfig.cc | |
| ms_to_VariantMatrix.cc | |
| msformat.hpp | |
| msstats.cc | |
| Mutation.hpp | |
| Mutation.tcc | |
| NeutralSample.hpp | |
| nsl.cc | |
| nSL.cc | |
| nsl.hpp | NSL and iHS |
| nSL.hpp | |
| nSL_from_ms.cc | |
| nvariablesites.cc | |
| nvariablesites.hpp | Calculate total numbers of polymorphisms |
| pairwiseld.cc | |
| PathwayHelper.cc | |
| PathwayHelper.hpp | Declarations of Sequence::Intermediates2 and Sequence::Intermediates3 |
| phylipData.hpp | Sequence::phylipData – read in phylip alignments |
| phylipData.tcc | |
| PolyFunctional.hpp | A collection of function objects for SNP analysis |
| PolySIM.cc | |
| PolySIM.hpp | Declaration of Sequence::PolySIM, a class to analyze coalescent simulation data |
| PolySIMtest.cc | |
| PolySites.cc | |
| PolySites.hpp | Sequence::PolySites, generates polymorphism tables from data |
| PolySites.tcc | |
| PolySitesIO.cc | |
| polySiteVector.cc | |
| polySiteVector.hpp | Site-major variation tables in ASCII format |
| polySiteVector_test.cc | Examples of using Sequence::polySiteVector |
| polySiteVectorTest.cc | Unit tests for Sequence::polySiteVector |
| PolySNP.cc | |
| PolySNP.hpp | Declaration of Sequence::PolySNP, a class to analyze SNP data |
| PolySNPimpl.hpp | |
| PolySNPtest.cc | |
| PolyTable.cc | |
| PolyTable.hpp | Sequence::PolyTable, a virtual base class for polymorphism tables |
| PolyTable.tcc | |
| PolyTableBadBehavior.cc | Unit test for really bad things |
| PolyTableConversions.cc | |
| PolyTableFunctions.cc | |
| PolyTableFunctions.hpp | Operations on non-const Sequence::PolyTable objects |
| PolyTableFunctions.tcc | |
| PolyTableIterators.cc | |
| PolyTableManip.cc | |
| PolyTableSlice.hpp | |
| PolyTableSlice.tcc | |
| PolyTableSliceTest.cc | Tests for Sequence/PolyTableSlice.hpp |
| PolyTableTweaking.cc | |
| Recombination.cc | |
| Coalescent/Recombination.hpp | |
| Recombination.hpp | Namespace Sequence::Recombination |
| Recombination.tcc | |
| RedundancyCom95.cc | |
| RedundancyCom95.hpp | Class Sequence::RedundancyCom95 |
| RedundancyCom95test.cc | |
| rmin.cc | |
| Seq.cc | |
| Seq.hpp | Class Sequence::Seq, an abstract base class for Sequences |
| Seq8test.cc | Unit tests for Sequence/Seq.hpp |
| SeqAlphabets.cc | |
| SeqAlphabets.hpp | |
| SeqConstants.cc | |
| SeqConstants.hpp | |
| SeqConversions.cc | |
| SeqEnums.hpp | Definition of enumeration types |
| SeqFunctors.hpp | |
| SeqProperties.hpp | |
| SeqRegexes.hpp | Various useful rexex-based functions for dealing with data. Declares Sequence::basic_dna_alphabet,Sequence::full_dna_alphabet, Sequence::pep_alphabet, and the function Sequence::validSeq |
| SeqUtilities.hpp | Declaration of Sequence::makeCountList (an alternative to Sequence::stateCounter), Sequence::internalGapCheck |
| shortestPath.cc | |
| shortestPath.hpp | Routines to find the shortest distance between any 2 codons, using Grantham's distance. Declares the class Sequence::shortestPath, and the functions Sequence::mutsShortestPath and Sequence::diffType |
| SimData.cc | |
| SimData.hpp | Declaration of Sequence::SimData, a class representing polymorphism data from coalescent simulations generated under an infinite-sites scheme |
| SimParams.cc | |
| SimParams.hpp | Sequence::SimParams reads in the parameters of Dick Hudon's coalescent simulation program. Used in conjunction with Sequence::SimData |
| SimpleSNP.cc | |
| SimpleSNP.hpp | Declaration of Sequence::SimpleSNP, a polymorphism table stream in a "spreadsheet" format |
| SimpleSNPIO.cc | |
| SimTypes.hpp | Declaration of types for coalescent simulation |
| SingleSub.cc | |
| SingleSub.hpp | Used by Sequence::Comeron95, class Sequence::SingleSub calculates divergence between codons that differ at one site |
| Sites.cc | |
| Sites.hpp | Class Sequence::Sites calculates the length of a pairwise alignment between coding sequences in terms of site degeneracy. Used by Sequence::Comeron95 |
| slidingWindow.cc | |
| slidingWindow2.cc | |
| Snn.cc | |
| Snn.hpp | |
| Snn.tcc | |
| Specializations.cc | Definitions of template specializations for library functions |
| stateCounter.cc | |
| stateCounter.hpp | Declaration of Sequence::stateCounter, a class to keep track of nucleotide counts either at a site in an alignment, or along a sequence |
| stateCounterTest.cc | |
| StateCounts.cc | |
| StateCounts.hpp | |
| SummStats.cc | |
| summstats.hpp | Include all summary statistic functions and types |
| SummStats.hpp | |
| tajd.cc | |
| test_SimDataIO.cc | |
| testAlleleCountMatrix.cc | Tests for Sequence/VariantMatrix.hpp |
| testClassicSummstats.cc | Unit tests for pop gen summary statistics |
| testClassicSummstatsEmptyVariantMatrix.cc | Unit tests for pop gen summary statistics |
| testGarudStatistics.cc | Unit tests for H1, H12, H2/H1 stats |
| testLD.cc | Unit tests for LD-related calculations |
| thetah.hpp | Fay and Wu's 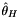 |
| thetah_thetal.cc | |
| thetal.hpp | Zeng et al. 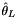 |
| thetapi.cc | |
| thetapi.hpp | |
| thetaw.cc | |
| thetaw.hpp | Watterson's theta |
| ThreeSubs.cc | |
| ThreeSubs.hpp | Used by Sequence::Comeron95, class Sequence::ThreeSubs calculates divergence between codons that differ at three sites |
| Trajectories.hpp | |
| Trajectories.tcc | |
| Translate.cc | |
| Translate.hpp | Declares Sequence::Translate,a function to translate CDS sequences into peptide sequences |
| Translate2.hpp | Deprecated header declaring routines to translate sequences. Including it includes the current header and issues a compiler warning |
| translateTest.cc | |
| TreeOperations.hpp | |
| TwoSubs.cc | |
| TwoSubs.hpp | Used by Sequence::Comeron95, class Sequence::TwoSubs calculates divergence between codons that differ at two sites |
| typedefs.hpp | Typedefs used by libsequence |
| ufs.cc | |
| Unweighted.cc | |
| Unweighted.hpp | Declares Sequence::Unweighted2 and Sequence::Unweighted3 |
| util.hpp | Helper functions for implementing summary statistics |
| valid_dna.cc | |
| variant_matrix_views_internal.hpp | |
| VariantMatrix.cc | |
| VariantMatrix.hpp | |
| VariantMatrixFixture.hpp | |
| VariantMatrixTest.cc | Tests for Sequence/VariantMatrix.hpp |
| VariantMatrixViews.cc | |
| VariantMatrixViews.hpp | |
| WeightingSchemes.hpp | Abstract interface to weighting schemes when codons differ at 2 positions |
| windows.cc | |
| windows.hpp |
